We are happy to share four new publications, published in the Proceedings of the 20th World Congress on Medical and Health Informatics. We have shown different applications of LLMs in healthcare:
- Large Language Models Reveal Menstruation Experiences and Needs on Social Media
- Enhancing Interpretability of Ocular Disease Diagnosis: A Zero-Shot Study of Multimodal Large Language Models
- Consensus Finding Among LLMs to Retrieve Information About Oncological Trials
- Large Language Models for Detection of Genetic Variants in Biomedical Literature
Large Language Models Reveal Menstruation Experiences and Needs on Social Media
Charlotte Tumescheit, Davinny Sou, Marcia Nißen, Tobias Kowatsch, Janna Hastings
The gender knowledge gap in medicine, particularly regarding menstruation and disorders such as endometriosis, often results in delayed diagnoses
and inadequate care. Many menstruating individuals report dismissal of debilitating
symptoms, driving them to seek information and support on online platforms such
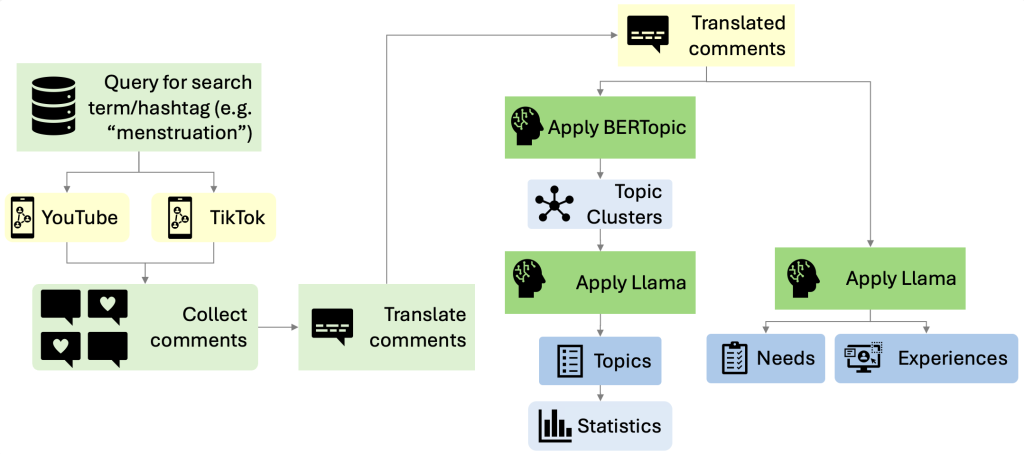
as TikTok and YouTube. This study leverages LLMs and social media to identify key topics reflecting lived experiences and needs to bridge this knowledge gap. Using a novel pipeline, we analysed video comments using BERTopic and the Llama 3.1 model.
Key topics, including emotional support, educational guidance, and community
validation, were consistent with prior research. This study underscores the potential
of social media and large language models to inform inclusive menstrual health
research, revealing unique insights regarding the menstruation experiences and
needs of underrepresented and historically overlooked individuals such as those
with irregular cycles.
Method overview:
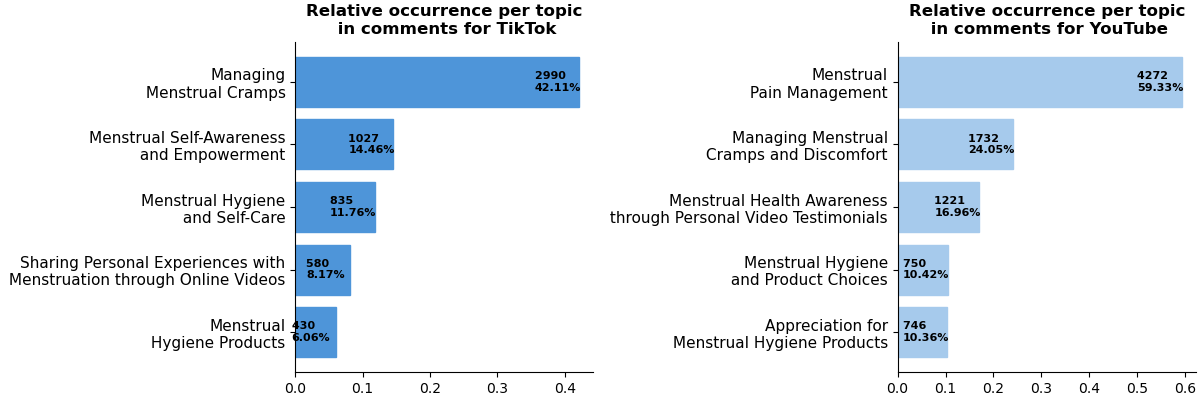
Selected results:
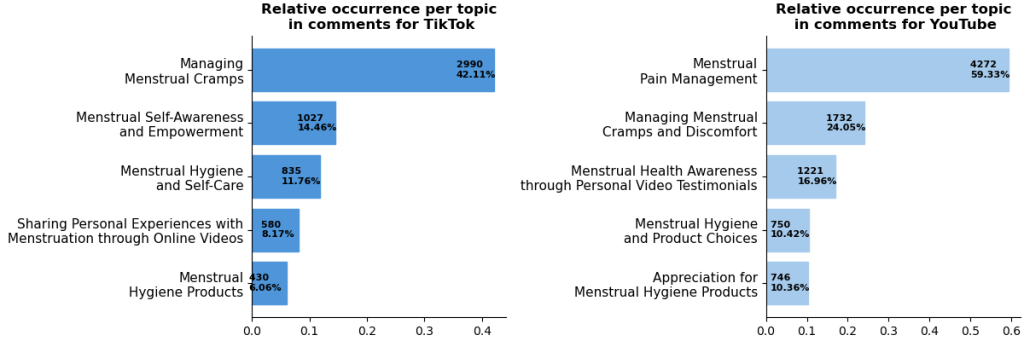
For more insights check out the paper: https://ebooks.iospress.nl/volumearticle/73968.
Enhancing Interpretability of Ocular Disease Diagnosis: A Zero-Shot Study of Multimodal Large Language Models
Yating Pan, Janna Hastings
Visual foundation models have advanced ocular disease diagnosis, yet
providing interpretable explanations remains challenging. We evaluate multimodal
LLMs for generating explanations of ocular diagnoses, combining Vision
Transformer-derived saliency maps with clinical metadata. After finetuning
RETFound for improved performance on the BRSET dataset (AUC-ROC
0.9664/0.8611 for diabetic retinopathy/glaucoma), we compared five LLMs
through technical and clinical evaluations. GPT-o1 demonstrated superior
performance across technical dimensions and clinical metrics (79.32% precision,
77.18% recall, 78.25% F1, 20.68% hallucination rate). Our findings highlight the
importance of underlying diagnostic accuracy and advanced model architecture for
generating reliable clinical explanations, suggesting opportunities for integrated
verification mechanisms in future developments. The code and details can be
found at: https://github.com/YatingPan/ocular-llm-explainability.
Selected results: Mean scores for five LLMs across five technical dimensions.
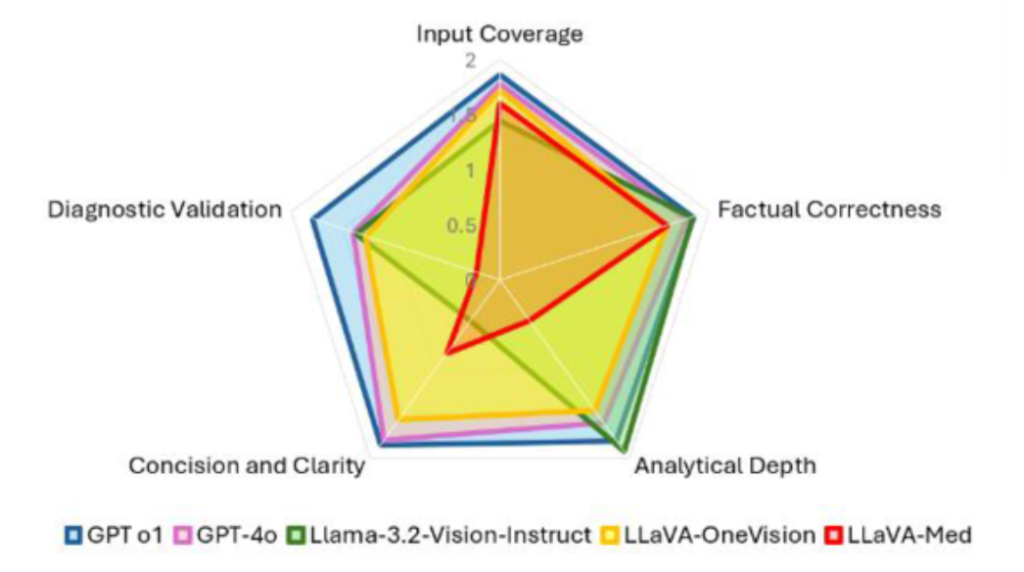
For more insights check out the paper: https://ebooks.iospress.nl/volumearticle/73938.
Consensus Finding Among LLMs to Retrieve Information About Oncological Trials
Fabio Dennstädt, Paul Windisch, Irina Filchenko, Johannes Zink, Paul Martin Putora, Ahmed Shaheen, Roberto Gaio, Nikola Cihoric, Marie Wosny, Stefanie Aeppli, Max Schmerder, Mohamed Shelan, Janna Hastings
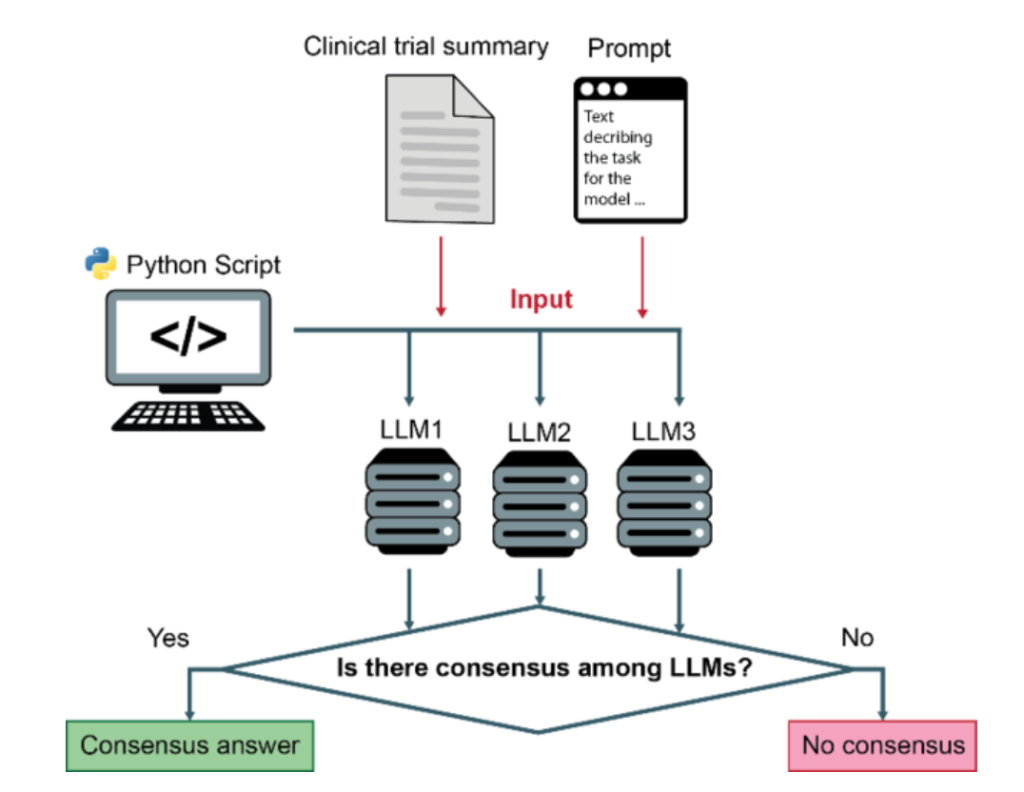
Background: Automated classification of medical literature is increasingly vital, especially in oncology. As shown in previous work, LLMs can be used as part of a flexible framework to accurately classify biomedical literature and trials. In the present study, we aimed to explore to what extent a consensus-based approach could improve classification performance.
Methods: The three LLMs Mixtral-8x7B, Meta-Llama-3.1-70B, and Qwen2.5-72B were used to classify oncological trials across four data sets with nine questions. Metrics (accuracy, precision, recall, F1-score) were assessed for individual models and consensus results.
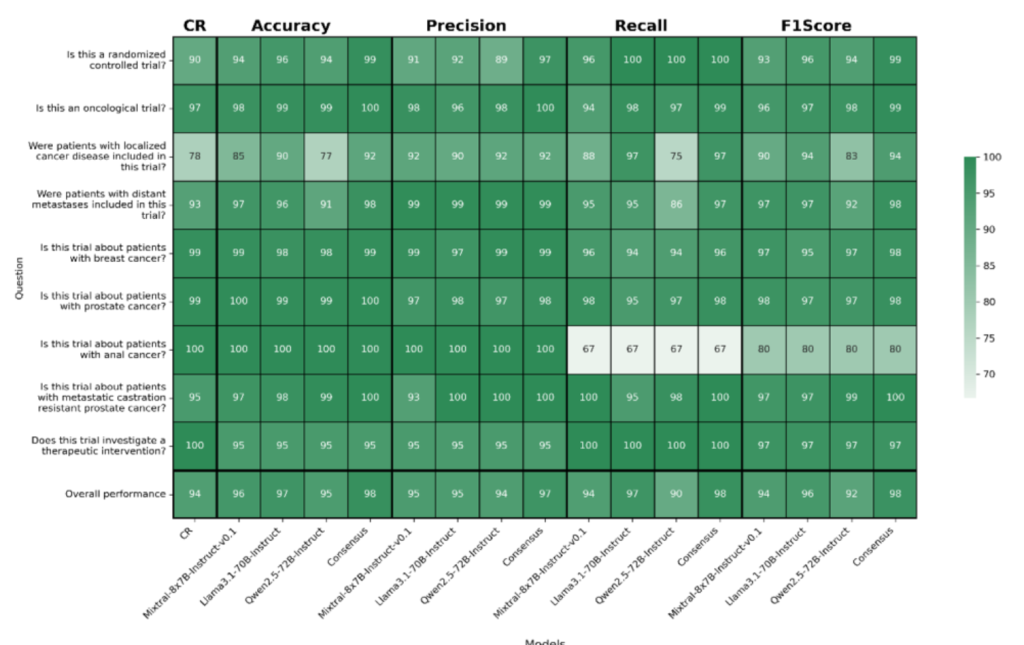
Results: Consensus was achieved in 93.93% of cases, improving accuracy
(98.34%), precision (97.01%), recall (98.11%), and F1-score (97.55%) over
individual models.
Conclusions: The consensus-based LLM framework delivers
high accuracy and adaptability for classifying oncological trials, with potential
applications in biomedical research and trial management.
Method overview:
Selected results: Classification metrics on different questions; Results in percentage; CR: Consensus rate
For more insights check out the paper: https://ebooks.iospress.nl/volumearticle/73865.
Large Language Models for Detection of Genetic Variants in Biomedical Literature
Marie Wosny, Janna Hastings
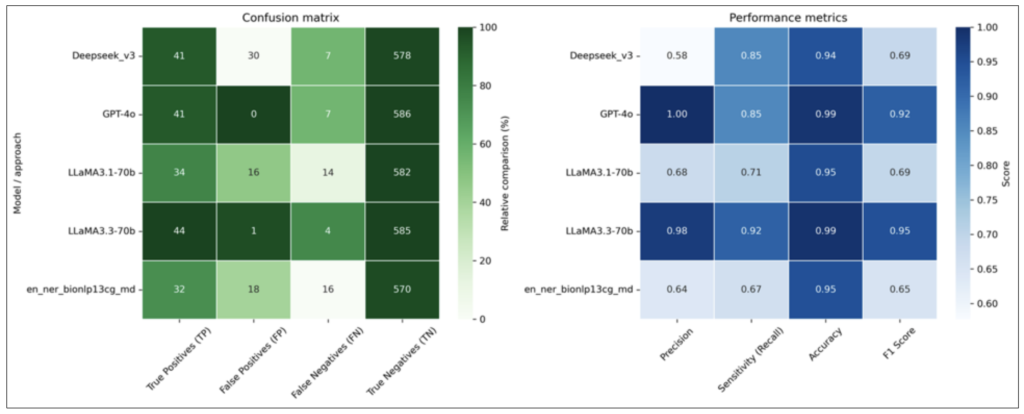
Personalized medicine relies on understanding genetic variations, but systematically tracking literature linking these variations to clinical outcomes remains challenging. This study assessed the feasibility and performance of large language models (LLMs) in extracting genes and variants from prostate cancer literature. Four LLMs, including LLaMA 3.3, GPT-4o, and DeepSeek-V3, were evaluated, with LLaMA-3.3-70b demonstrating the highest overall performance (98% precision, 92% recall, 99% accuracy). In contrast, conventional information extraction methods had poor recall and high false positive rates. Moreover, LLMs inferred contextual details, offering enriched insights but occasionally introducing unsupported information. These findings demonstrate the promise of LLMs in automating genomic variant extraction, while also highlighting the need for rigorous validation and careful integration into clinical and research workflows.
Selected results: Confusion matrix and performance metrics for variant detection models and approaches.
For more insights check out the paper: https://ebooks.iospress.nl/volumearticle/73864.